Bacterial populations inhabiting ultrapure water (UPW) systems were investigated. The analyzed UPW systems included pilot scale, bench scale, and full size UPW plants employed in the semiconductor and other industries. Bacteria present in the polishing loop of the UPW systems were enumerated by both plate counts and epifluorescence microscopy. Assessment of bacterial presence in UPW by epifluorescence microscopy (cyanotolyl tetrazolium chloride [CTC] and DAPI staining) showed signifi- cantly higher numbers (10 to 100 times more bacterial cells were detected) than that determined by plate counts. A considerable proportion of the bacteria present in UPW (50 to 90%) were cells that did not give a positive signal with CTC stain. Bacteria isolated from the UPW systems were mostly gram negative, and several groups seem to be indigenous for all of the UPW production systems studied. These included Ralstonia pickettii, Bradyrhizobium sp., Pseudomonas saccharophilia, and Stenotrophomonas strains. These bacteria constituted a significant part of the total number of isolated strains (>20%). Two sets of primers specific to R. pickettii and Bradyrhizobium sp. were designed and successfully used for the detection of the corresponding bacteria in the concentrated UPW samples. Unexpectedly, nifH gene sequences were found in Bradyrhizobium sp. and some P. saccharophilia strains isolated from UPW. The widespread use of nitrogen gas in UPW plants may be associated with the presence of nitrogen-fixing genes in these bacteria.
Many industries suffer from the microbial contamination of ultrapure water (UPW). These include the semiconductor, pharmaceutical, food, and beverage industries. Within the semiconductor industry, ultrapure water is utilized in the final rinsing stage, and the presence of even a single bacterial cell and/or the products of cellular degradation, can severely compromise the quality of the final product.
The industrial production of UPW is a complex multistep process, which involves two major stages referred to as pretreatment and polishing (Fig. 1). A variety of steps are included in many UPW production systems (e.g., filtration, UV light treatment, heat treatment, and ozonation) to remove and destroy bacteria. In particular, treatment with UV254 light and ozonation are present in some parts of a facility solely to prevent microbial contamination. Nitrogen gas is often used instead of air above stored UPW to prevent carbon dioxide and oxygen from dissolving in the water. It is imperative that UPW is kept carbon dioxide-free to prevent ionic loading on the mixed-bed ion-exchange resins, while the lowering of oxygen concentration should minimize bacterial growth (Fig. 1). Despite these precautions, piping, membranes, tanks, and other surfaces within the UPW system provide favorable places for bacterial adhesion and cell growth. The complete removal of contaminating microorganisms is considered to be nearly impossible.
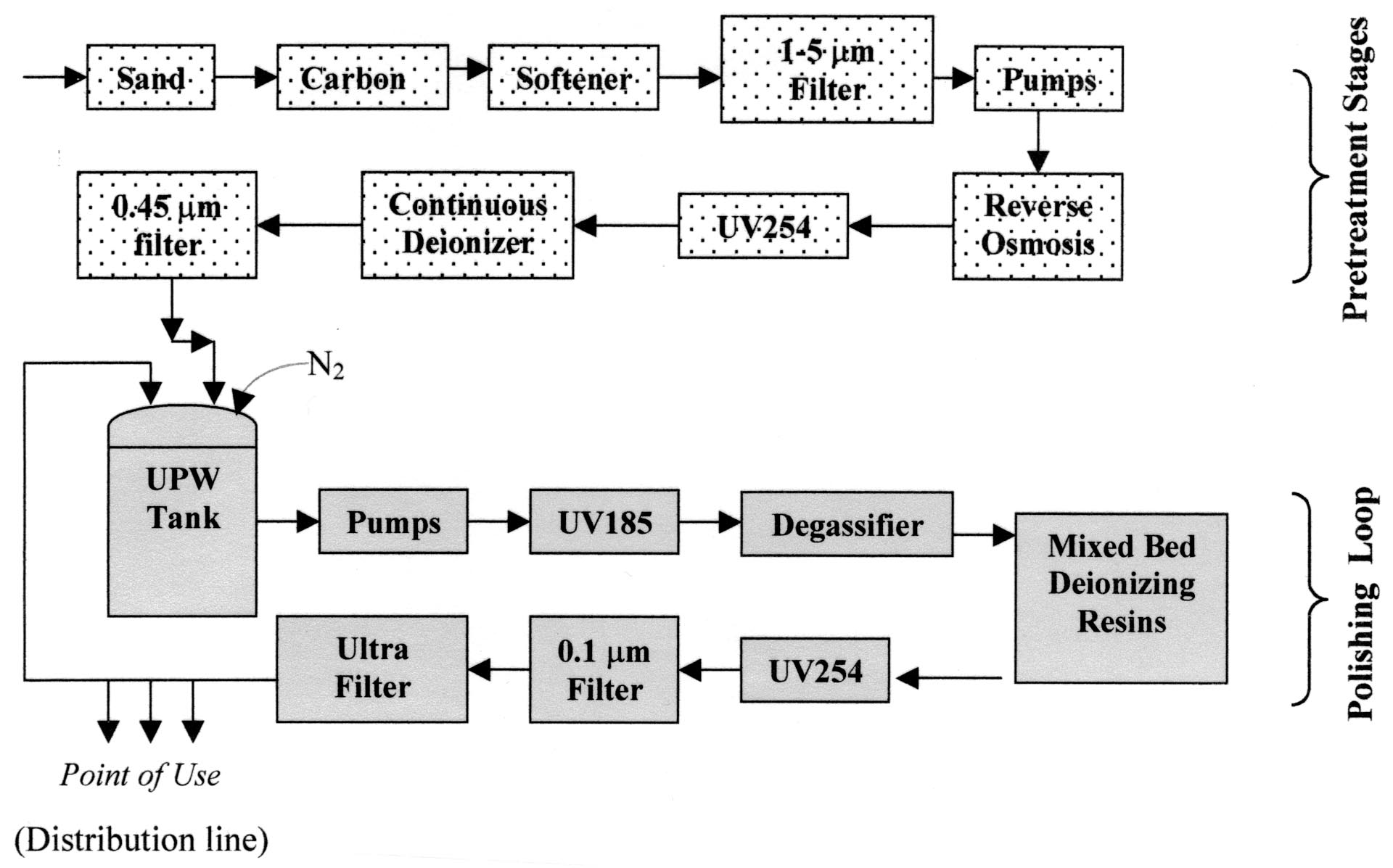
Fig1
We investigated here the diversity of bacterial communities in two university and four industrial UPW systems. More emphasis was placed on the microorganisms found in the polishing loops (especially distribution lines) of the systems. Oligonucleotide probes specific to the main bacterial species inhabiting UPW were designed. These probes were then successfully used to directly detect bacteria present in five different UPW plants. Pseudomonas, Ralstonia, and Bradyrhizobium species were shown to be present in most of the analyzed UPW systems.
MATERIALS AND METHODS Media and bacterial strains. R2A media (28) was used in this work for growth and analysis of the bacterial strains present in UPW. This medium is recommended by American Society for Testing and Materials (ASTM) (1, 2) for testing UPW quality and is therefore widely used in industries. All bacterial strains investigated in this study were isolated from water samples obtained at different UPW plants. Three of the six plants analyzed in this study are used for production of UPW in semiconductor manufacturing processes. Designation of the UPW systems analyzed in this work is as follows: UPWS-1 (University of Arizona experimental UPW system, pilot scale), UPWS-2 (University of Arizona experimental UPW system number 2, bench scale), UPWS-3 (industrial UPW system number 1), UPWS-4 (industrial UPW system number 2), UPWS-5 (industrial UPW system number 3, not semiconductor industry), and UPWS-6 (industrial UPW system number 3, not semiconductor industry). Figure 1 shows a schematic of the common parts of the analyzed UPW systems. Nitrogen was used in polishing loops of UPWS-1, UPWS-3, UPWS-5, and UPWS-6.
For detection of 16S rRNA gene sequences in UPW by PCR (direct PCR) bacterial cells from the UPW were concentrated onto polycarbonate membranes (0.1-μm pore size) as described above. The membrane was aseptically removed from the filter holder and transferred to a sterile polypropylene centrifuge tube (50 ml) containing 10 ml of double-filter-sterilized UPW. This was incubated at 25°C (180 rpm) overnight. After incubation, the cells were concentrated by centrifugation (8,000 rpm, 15 min) and resuspended in 1 ml of double-filtersterilized UPW. Concentrated water samples were used directly in the PCRs.
Alignments of the sequences were performed by using the CLUSTALW program. Phylogenetic analysis of the alignment was done by using the PHYLIP (version 3.57c) package (10) and the TREECON program (34). For the PHYLIP analysis, bootstraps were obtained with the SEQBOOT program (100 data sets were generated). Parsimony analyses were done with the DNAPARS programs with ordinary parsimony and randomized input order of the sequences. For the analyses with the TREECON program, Tajima and Nei correction (31) was used, and trees were generated by neighbor joining.
After initial characterization of the isolated strains, corresponding 16S rRNA gene sequences were obtained. Sequences of at least 900 bp were determined, and for every group of closely related sequences (homology of >99%), an almost complete 16S rRNA gene sequence (1,400 to 1,500 bp) was obtained for at least one bacterial isolate. The results of phylogenetic analysis of bacteria present in UPW obtained from five different plants are shown in Table 1, and a phylogenetic tree of the main bacterial strains isolated from UPW (UPWS- 1) is presented in Fig. 2. All of the UPW samples analyzed were collected from ports situated in the final (polishing loop) parts of the corresponding UPW systems (mostly the distribution line, indicated in Fig. 1).
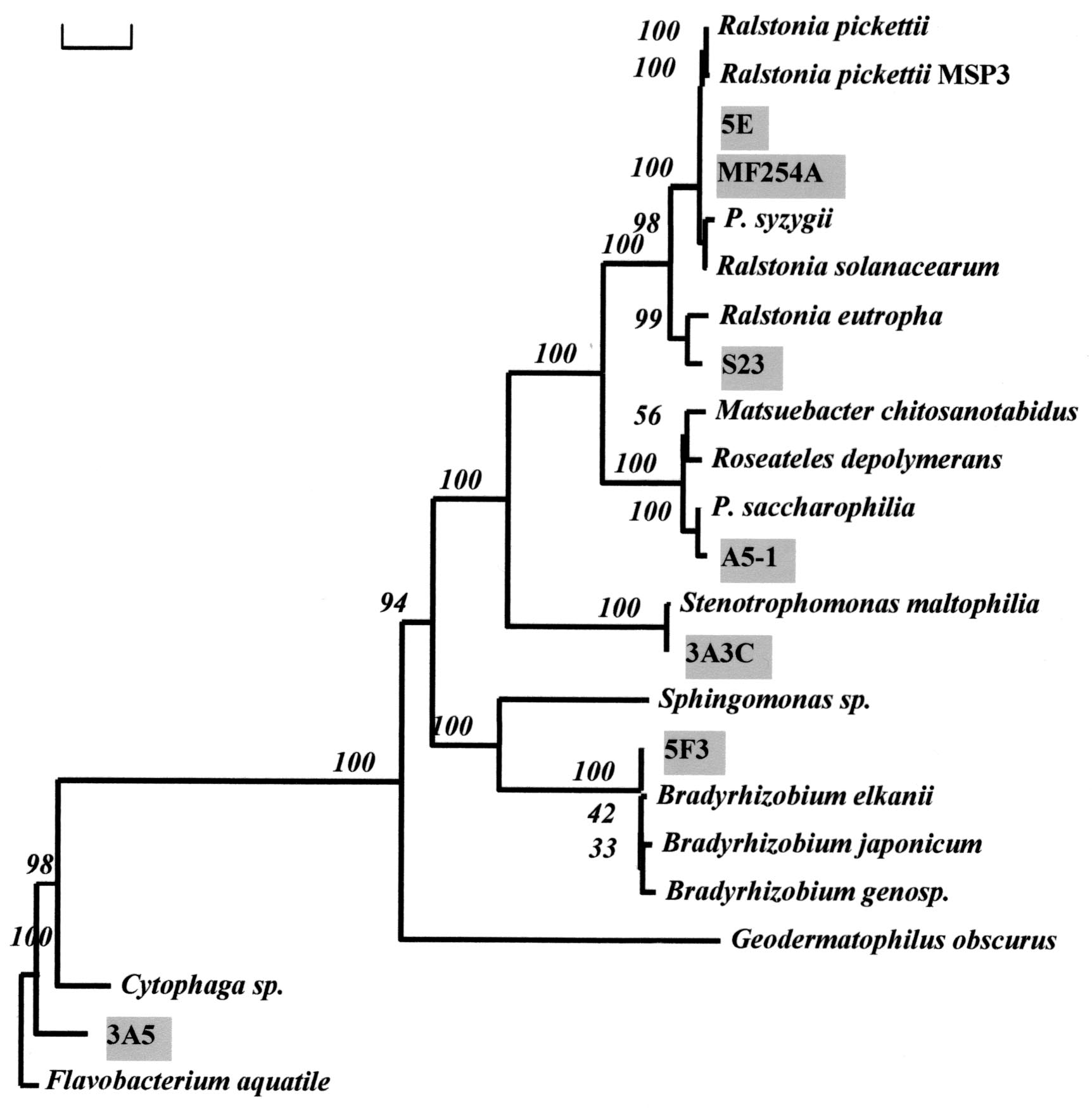
Fig2
The bacterial diversity within the UPW systems primarily employed in the semiconductor industry has been investigated. Six UPW systems were analyzed, two smaller university systems and four full-size industrial plants, that were located in geographically diverse areas of the world. Because the UPW systems varied in size and location, the results obtained are considered to be characteristic for UPW production in general. The samples analyzed here were obtained from the ports located in the polishing sections of UPW systems; hence, the bacterial communities investigated may have a significant impact on the quality of UPW used in the final (rinsing) stages of semiconductor production.
Discovery of nifH genes in Bradyrhizobium strains is not surprising by itself, but when taken in conjunction with the spread of this bacterial group in UPW systems, it may suggest that nitrogen contributes to cell maintenance and growth in these systems. The presence of nifH sequences in P. saccharophilia is somewhat more unusual. However, a few examples of nitrogen-fixing Pseudomonas strains have been reported, and a nitrogen-fixing strain of P. saccharophilia ATCC 15946 has also been reported (3). It is important to stress that finding bacterial strains with nif genes deserves further investigation, since it provides the first evidence that the widespread use of nitrogen in UPW production systems may contribute to bacterial contamination.
Identification of the typical bacterial strains inhabiting UPW systems allowed us to design oligonucleotide primers specific to two main bacterial groups. PCR experiments conducted with UPW samples demonstrated the possibility for detection of R. pickettii and Bradyrhizobium sp. in UPW. A 100- to 1,000- fold concentration of water samples was needed for such detection, since bacterial numbers in typical UPW taken from the system were too low to allow direct PCR detection of bacterial 16S rDNA sequences. The use of specific (as well as universal) primers for the detection of the bacterial contamination in UPW systems may be considered useful for the preliminary assessment of bacterial presence in UPW.
上一篇: 微孔膜组件去除超纯水生产中的溶解氧
下一篇: 应用于半导体制造的超纯水工艺